Recombinant Saccharomyces cerevisiae Eukaryotic peptide chain release factor GTP-binding subunit (SUP35), partial
-
中文名称:酿酒酵母SUP35重组蛋白
-
货号:CSB-YP356520SVG
-
规格:
-
来源:Yeast
-
其他:
-
中文名称:酿酒酵母SUP35重组蛋白
-
货号:CSB-EP356520SVG
-
规格:
-
来源:E.coli
-
其他:
-
中文名称:酿酒酵母SUP35重组蛋白
-
货号:CSB-EP356520SVG-B
-
规格:
-
来源:E.coli
-
共轭:Avi-tag Biotinylated
E. coli biotin ligase (BirA) is highly specific in covalently attaching biotin to the 15 amino acid AviTag peptide. This recombinant protein was biotinylated in vivo by AviTag-BirA technology, which method is BriA catalyzes amide linkage between the biotin and the specific lysine of the AviTag.
-
其他:
-
中文名称:酿酒酵母SUP35重组蛋白
-
货号:CSB-BP356520SVG
-
规格:
-
来源:Baculovirus
-
其他:
-
中文名称:酿酒酵母SUP35重组蛋白
-
货号:CSB-MP356520SVG
-
规格:
-
来源:Mammalian cell
-
其他:
产品详情
-
纯度:>85% (SDS-PAGE)
-
基因名:SUP35
-
Uniprot No.:
-
别名:SUP35; GST1; PNM2; SAL3; SUF12; SUP2; YDR172W; YD9395.05; Eukaryotic peptide chain release factor GTP-binding subunit; ERF-3; ERF3; ERF2; G1 to S phase transition protein 1; Omnipotent suppressor protein 2; PSI no more protein 2; Polypeptide release factor 3; Translation release factor 3
-
种属:Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast)
-
蛋白长度:Partial
-
蛋白标签:Tag type will be determined during the manufacturing process.
The tag type will be determined during production process. If you have specified tag type, please tell us and we will develop the specified tag preferentially. -
产品提供形式:Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
复溶:We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20℃/-80℃. Our default final concentration of glycerol is 50%. Customers could use it as reference.
-
储存条件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保质期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
货期:Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.Note: All of our proteins are default shipped with normal blue ice packs, if you request to ship with dry ice, please communicate with us in advance and extra fees will be charged.
-
注意事项:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet :Please contact us to get it.
问答及客户评论
Could you pls let us know the species ?
Here is the reference link: http://www.uniprot.org/uniprot/?query=Sup35&sort=score
Could you pls help to confirm ?
Is it convenient to provide the sequence or Uniprot No + expression region ?
靶点详情
-
功能:Involved in translation termination. Stimulates the activity of ERF1. Binds guanine nucleotides. Recruited by polyadenylate-binding protein PAB1 to poly(A)-tails of mRNAs. Interaction with PAB1 is also required for regulation of normal mRNA decay through translation termination-coupled poly(A) shortening.
-
基因功能参考文献:
- The role of SUP35 in protein folding in the ribosome and prion propagation PMID: 27633137
- With the use of both segmental and specific isotopic labeling schemes in combination with dynamic nuclear polarization (DNP) NMR, the study examined an amyloid form of Sup35NM that does not have a parallel in-register structure. PMID: 28330994
- In stressed conditions, Sup35 formed protective gels via pH-regulated liquid-like phase separation followed by gelation. PMID: 29301985
- proteolytic cleavage of Sup35 suppresses spontaneous de novo generation of the [PSI(+)] prion. PMID: 29038292
- we discovered a non-chromosomal factor, [NSI+], which possesses the main features of yeast prions, including cytoplasmic infectivity, reversible curability, dominance, and non-Mendelian inheritance in meiosis. This factor causes omnipotent suppression of nonsense mutations in strains of S. cerevisiae bearing a deleted or modified Sup35 N-terminal domain PMID: 28027291
- our observations suggest that these sequence elements make complementary contributions to prion propagation, with the repeat proximal region promoting chaperone binding to and the repeats promoting chaperone processing of Sup35 amyloid PMID: 27814358
- Sup35 becomes oxidized and aggregates during oxidative stress conditions and plays a role in [PSI+] prion formation. PMID: 28369054
- It was found that ABF1, FKH2, and REB1 overexpression decreased the growth of strains in a prion-dependent manner and, thus, might influence [PSI;(+)] prion toxicity. PMID: 27830682
- Autophagy therefore normally functions to remove oxidatively damaged Sup35, which accumulates in cells grown under aerobic conditions, but in the absence of autophagy, damaged/misfolded Sup35 undergoes structural transitions . PMID: 26490118
- DnaJB6 prevented purified Sup35NM from forming amyloids at 37 degrees C, which produce predominantly weak [PSI(+)] variants when used to infect yeast PMID: 26702057
- findings show Sup35p is secreted within extracellular vesicles (EV)released in the extracellular medium of yeast cultures; demonstrated that Sup35p within EV isolated from strong and weak [PSI(+)] cells is in an infectious prion conformation PMID: 26286691
- Dynamic nuclear polarization of nuclear magnetic resonance of Sup35 amyloid fibers, NM, show structure in its biological context indicates that these organizing protein-protein interactions are mediated through the M domain of the protein via the adoption of a beta sheet secondary structure by the majority of this otherwise intrinsically disordered region. PMID: 26456111
- findings show that oxidation of Sup35 is a common response to oxidative stress and results in Sup35 conversion to the [PSI+] state PMID: 25601439
- A hybrid modelling approach is presented enabling detailed analysis of a highly ordered and hierarchically organized fibril of the GNNQQNY peptide fragment of a yeast prion protein SUP35. PMID: 25849399
- Amino Acid Proximities in Two Sup35 Prion Strains PMID: 26265470
- Data indicate mutations decrease amyloid formation in the prion protein Sup35. PMID: 24817093
- The data obtained have refined a supposed organization of beta-sheets forming by different regions of Sup35p prion-forming domain within amyloid. PMID: 25850301
- Sup35(7-13) is observed to organize into anti-parallel and parallel beta-sheet arrangements. Confinement in the sodium bis(2-ethylhexyl) sulfosuccinate reverse micelles stabilizes extended peptide conformations and enhance peptide aggregation. PMID: 25494801
- [PIN+] drives Pin4C into a toxic hyperphosphorylated species. PMID: 25039275
- analysis of amino acid requirements for formation and maintenance of the [PSI] prion in yeast PMID: 25547291
- In Saccharomyces cerevisiae the association of Pab1 with eRF3 negatively regulates translation termination. PMID: 25411355
- the influence of SUP35 and VTS1 on [NSI+] PMID: 25474892
- Strikingly, exposure to specific stressful environments dramatically altered the variants of Sup35 [PSI(+)] that formed de novo. PMID: 24628771
- Lsb1 is crucial for prion maintenance during stress. PMID: 25143386
- Sup35pC may serve as actin modulator during mitosis. PMID: 24549315
- Sup35p is degraded in vitro by the 26S and 20S proteasomes in a ubiquitin-independent manner, generating an array of amyloidogenic peptides derived from its prion-domain. PMID: 24589377
- Data shows that the translation termination factor Sup35 is required for premature translation termination. PMID: 25154418
- The Sup35/Pub1 complex, which also contains TUB1 mRNA and components of translation machinery, is important for the integrity of the tubulin cytoskeleton. PMID: 24981173
- Data indicate that the Sup35(GPI) aggregates corresponded to dense cell surface accumulations of membrane vesicle-like structures and were not fibrillar. PMID: 24627481
- A conformation analysis of both full-length Sup35 and also from its isolated NM domain shows that Sup35NM and fragments thereof, which are often used as convenient models for prion fibril assembly, have a very different conformation of the prion domains. PMID: 24123863
- Authors show that even within a single cell, Sup35 retains the potential to fold into more than one variant type. PMID: 22998111
- the effect of Hofmeister ions on amyloid nucleation and strain generation by the prion domain-containing fragment (Sup35NM) of a yeast protein Sup35p. PMID: 23990463
- Data indicate that SUP35 ORF (eRF3) affects binding between SUP45 ORF (eRF1) and the ribosome, either prior to or subsequent to peptide release. PMID: 23963452
- This study reveals an unexpected interplay between the wild type Sup35p and proteins expressed from the sup35(KK) alleles during prionization. PMID: 23965990
- found that over-production of C-terminally truncated multi-transmembrane (MTM) mutant proteins triggers HSR, but not UPR, and clearance of yeast prions [PSI(+)] and [URE3] PMID: 23384482
- Mutations in a Gly58-Gly59 pair in Sup35 inhibit propagation of the [PSI+] prion. PMID: 23746351
- We find that among the rare wild isolates containing [PSI(+)], all indistinguishably "weak" [PSI(+)], are several different variants based on their transmission efficiencies to other Sup35 alleles PMID: 23382698
- The yeast prions [URE3] and [PSI] are not found in wild strains, suggesting they are not an advantage.Recently, we showed that the array of [PSI] and [URE3] prions includes a majority of lethal or very toxic variants. PMID: 22052353
- Study investigated the interaction between Hsp104 and Sup35, the priongenic protein in yeast that forms the [PSI+] prion; found that a 20-amino acid segment within the highly-charged, unstructured middle domain of Sup35 contributes to the physical interaction between the middle domain and Hsp104. PMID: 22561166
- Data indicate that the toxicity of 103 glutamines (HttQ103) in yeast containing the [PSI(+)] prion is primarily due to sequestration of the essential protein, Sup35. PMID: 22573320
- genetic interaction is present between SKY1 and SUP35 in the presence of diamide and hydrogen peroxide in the BY background but not in the RM background. PMID: 22396662
- SUP35 gene modulates nonsense suppression in S. cerevisiae PMID: 22215010
- a causal link between Sup35 protein oxidation and de novo [PSI(+)] prion formation. PMID: 21832086
- Data show that full-length New1 broke the Sup35NM amyloid fibrils in an ATP-dependent manner. PMID: 21453424
- Class I deletions (bug1Delta, bem1Delta, arf1Delta, and hog1Delta) reduced the efficiency of [PSI+] induction, but formed rings normally. Class II deletions (las17Delta, vps5Delta, and sac6Delta) inhibited both [PSI+] induction and ring formation PMID: 21625618
- Data report here over half of Saccharomyces cerevisiae [PSI(+)] variants are sick or lethal, and give a more realistic picture of the impact of the prion change than does focus on "mild" prion variants. PMID: 21402947
- By negative staining, cryo-EM and scanning transmission EM (STEM), filaments of full-length Sup35p show a thin backbone fibril surrounded by a diffuse 65-nm-wide cloud of globular C-domains. PMID: 21219467
- aggregation of Sup35p strictly depended on the presence of MgCl(2) stress in our strains PMID: 20937039
- show that the yeast Tsa1/Tsa2 peroxiredoxins colocalize to ribosomes and function to protect the Sup35 translation termination factor against oxidative stress-induced formation of its heritable [PSI(+)] prion conformation. PMID: 20308573
- sup35 mutations do not only decrease translation fidelity but also influence mRNA stability. PMID: 20198859
显示更多
收起更多
-
亚细胞定位:Cytoplasm.
-
蛋白家族:TRAFAC class translation factor GTPase superfamily, Classic translation factor GTPase family, ERF3 subfamily
-
数据库链接:
KEGG: sce:YDR172W
STRING: 4932.YDR172W
Most popular with customers
-
Express system: Mammalian cell
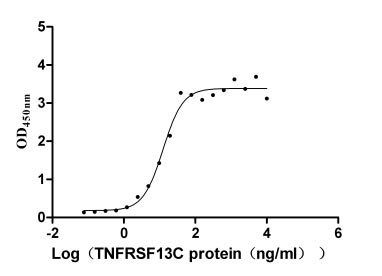
Species: Homo sapiens (Human)
-
Recombinant Human Delta-like protein 3 (DLL3), partial (Active)
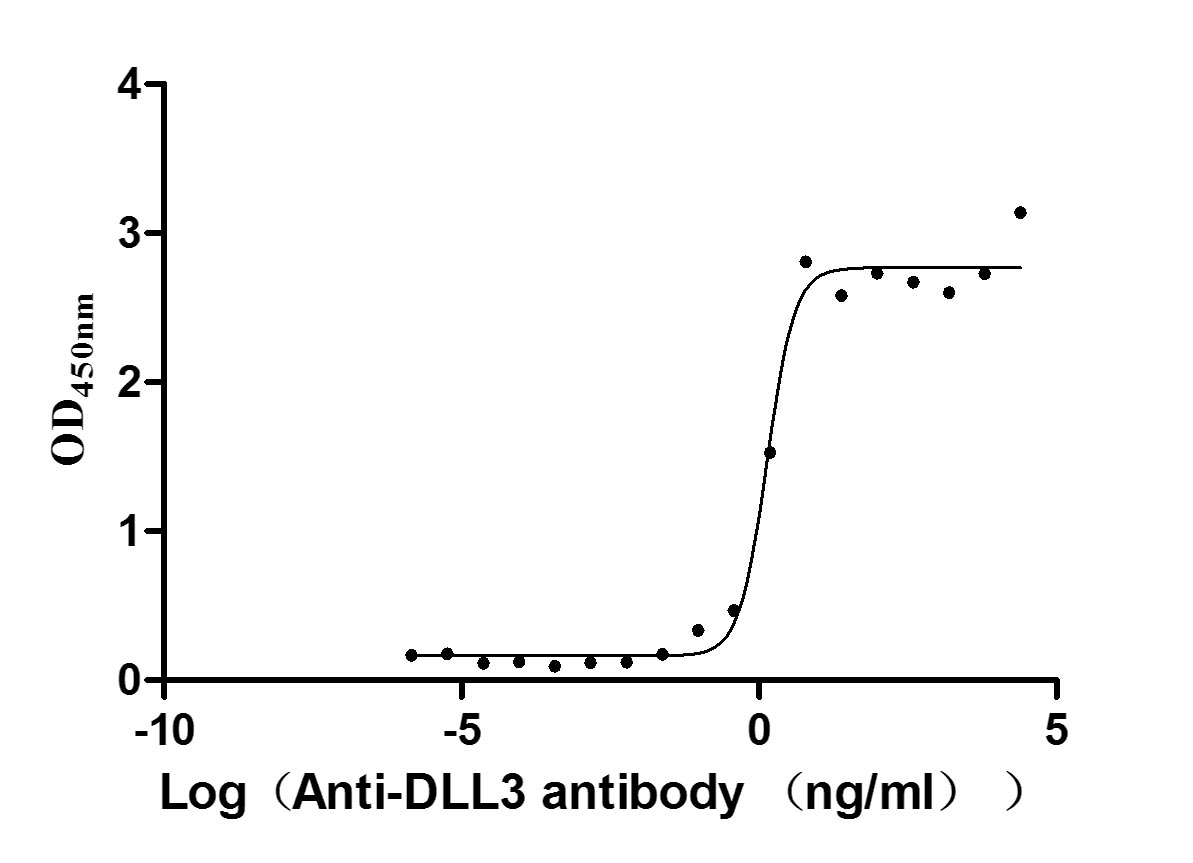
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
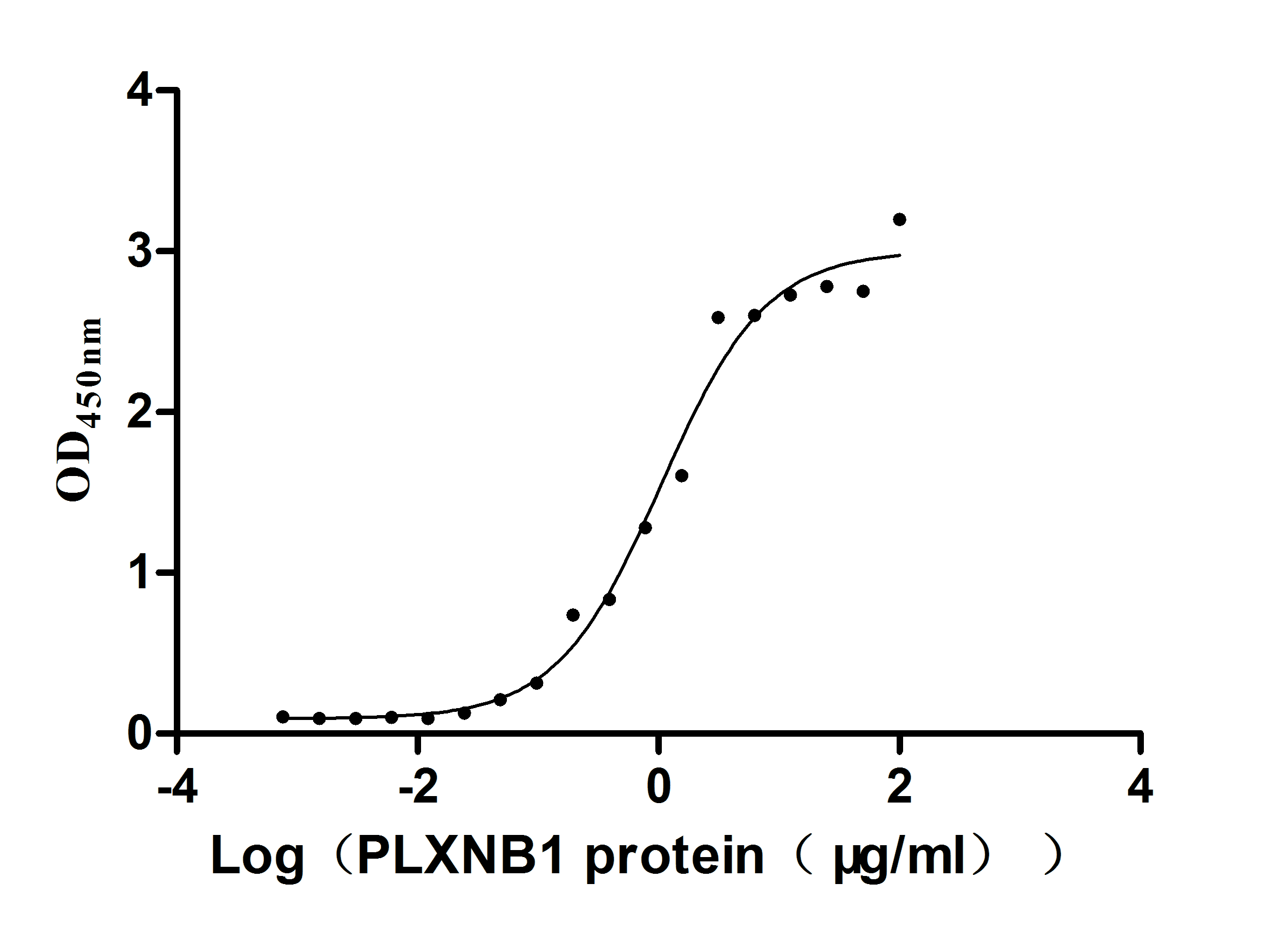
Recombinant Human Plexin-B1 (PLXNB1), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
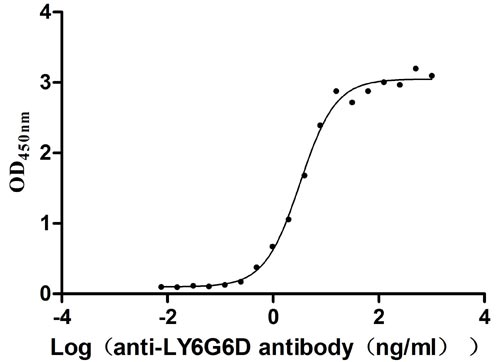
-
Recombinant Human Lymphocyte antigen 6 complex locus protein G6d (LY6G6D) (Active)
Express system: Yeast
Species: Homo sapiens (Human)
-
Recombinant Mouse Cell adhesion molecule 1 (Cadm1), partial (Active)
Express system: Mammalian cell
Species: Mus musculus (Mouse)
-
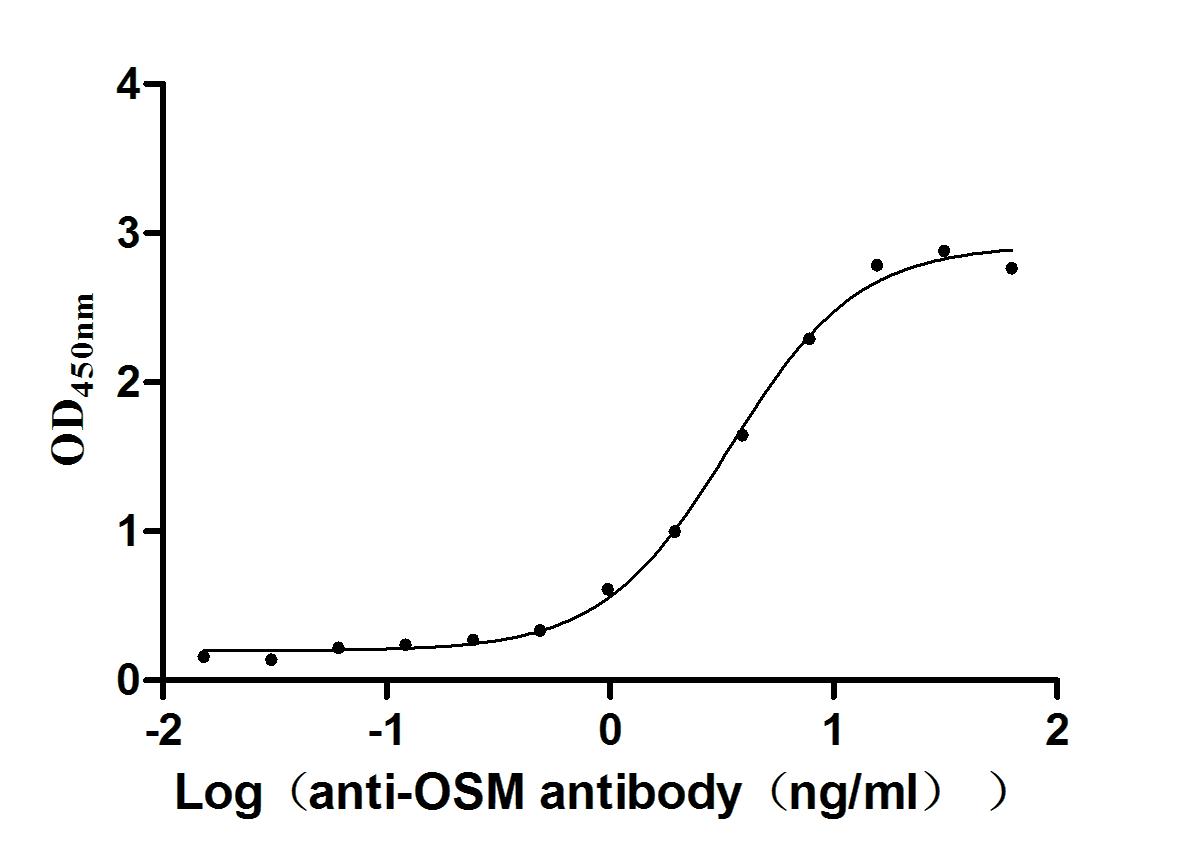
Recombinant Human Oncostatin-M (OSM), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
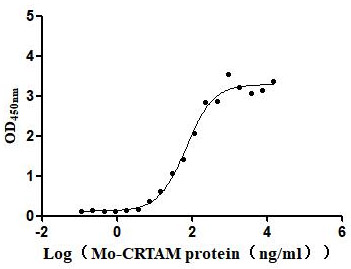
Recombinant Mouse Cytotoxic and regulatory T-cell molecule (Crtam), partial (Active)
Express system: Mammalian cell
Species: Mus musculus (Mouse)
-
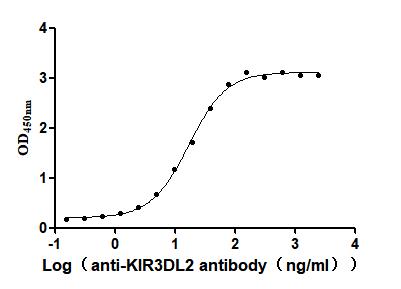
Recombinant Human Killer cell immunoglobulin-like receptor 3DL2 (KIR3DL2), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)