Recombinant Human Structure-specific endonuclease subunit SLX4 (SLX4), partial
-
中文名称:人SLX4重组蛋白
-
货号:CSB-YP810299HU
-
规格:
-
来源:Yeast
-
其他:
-
中文名称:人SLX4重组蛋白
-
货号:CSB-EP810299HU
-
规格:
-
来源:E.coli
-
其他:
-
中文名称:人SLX4重组蛋白
-
货号:CSB-EP810299HU-B
-
规格:
-
来源:E.coli
-
共轭:Avi-tag Biotinylated
E. coli biotin ligase (BirA) is highly specific in covalently attaching biotin to the 15 amino acid AviTag peptide. This recombinant protein was biotinylated in vivo by AviTag-BirA technology, which method is BriA catalyzes amide linkage between the biotin and the specific lysine of the AviTag.
-
其他:
-
中文名称:人SLX4重组蛋白
-
货号:CSB-BP810299HU
-
规格:
-
来源:Baculovirus
-
其他:
-
中文名称:人SLX4重组蛋白
-
货号:CSB-MP810299HU
-
规格:
-
来源:Mammalian cell
-
其他:
产品详情
-
纯度:>85% (SDS-PAGE)
-
基因名:SLX4
-
Uniprot No.:
-
别名:BTB (POZ) domain containing 12; BTB/POZ domain-containing protein 12; BTBD12; FANCP; KIAA1784; KIAA1987; MUS312; SLX 4; slx4; SLX4 structure-specific endonuclease subunit homolog (S. cerevisiae); SLX4_HUMAN; Structure-specific endonuclease subunit slx4
-
种属:Homo sapiens (Human)
-
蛋白长度:Partial
-
蛋白标签:Tag type will be determined during the manufacturing process.
The tag type will be determined during production process. If you have specified tag type, please tell us and we will develop the specified tag preferentially. -
产品提供形式:Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
复溶:We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20℃/-80℃. Our default final concentration of glycerol is 50%. Customers could use it as reference.
-
储存条件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保质期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
货期:Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.Note: All of our proteins are default shipped with normal blue ice packs, if you request to ship with dry ice, please communicate with us in advance and extra fees will be charged.
-
注意事项:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet :Please contact us to get it.
靶点详情
-
功能:Regulatory subunit that interacts with and increases the activity of different structure-specific endonucleases. Has several distinct roles in protecting genome stability by resolving diverse forms of deleterious DNA structures originating from replication and recombination intermediates and from DNA damage. Component of the SLX1-SLX4 structure-specific endonuclease that resolves DNA secondary structures generated during DNA repair and recombination. Has endonuclease activity towards branched DNA substrates, introducing single-strand cuts in duplex DNA close to junctions with ss-DNA. Has a preference for 5'-flap structures, and promotes symmetrical cleavage of static and migrating Holliday junctions (HJs). Resolves HJs by generating two pairs of ligatable, nicked duplex products. Interacts with the structure-specific ERCC4-ERCC1 endonuclease and promotes the cleavage of bubble structures. Interacts with the structure-specific MUS81-EME1 endonuclease and promotes the cleavage of 3'-flap and replication fork-like structures. SLX4 is required for recovery from alkylation-induced DNA damage and is involved in the resolution of DNA double-strand breaks.
-
基因功能参考文献:
- observed aberrant bands of CSMD1 in one case of CRCs and SRPK1 in two cases of colorectal cancers PMID: 29258766
- Findings propose a risk variant with high penetrance on the haplotype spanning SLX4/FANCP to be functionally associated to BC predisposition via decreased repair capacity and suggest this variant is carried by a fraction of these haplotypes that is enriched in early onset BC cases. PMID: 29044504
- Loss-of-function mutations in SLX4 may contribute to the development of breast cancer in very rare cases PMID: 23840564
- The BLM-TOP3A-RMI (BTR) dissolvase complex is required for Alternative lengthening of telomeres-mediated telomere synthesis. BLM and SLX4 play opposing roles in recombination-dependent replication at human telomeres. PMID: 28877996
- These data also indicate that HIV-1 and HIV-2 Vpr activate the DNA damage response through an SLX4-independent mechanism that remains uncharacterized. PMID: 27624129
- The functioning of SLX4 is dependent on its dimerization via an oligomerization motif called the BTB domain. PMID: 27131364
- Results identified homozygous mutations in FANCA and FANCP/SLX4 genes, both located on chromosome 16, were the affected recessive FA genes in three and one family respectively. Genotyping with short tandem repeat markers and SNP arrays revealed uniparental disomy of the entire mutation-carrying chromosome 16 in all four patients. PMID: 26841305
- SLX4 (FANCP) and XPF (FANCQ) proteins interact with each other and play a vital role in the Fanconi anemia (FA) DNA repair pathway. Study has revealed that the global minor allele, SLX4(Y546C), is defective in this interaction. PMID: 26453996
- SUMOylation and PARylation cooperate to recruit and stabilize SLX4 at DNA damage sites. PMID: 25722289
- Identification and characterization of MUS81 point mutations that abolish interaction with the SLX4 scaffold protein. PMID: 25224045
- Vpr recruits the SLX4 endonuclease complex and Vpr-induced inappropriate activation of this complex leads to cell cycle arrest at the G2 phase. PMID: 25496524
- FANCP has versatile functions in genome maintenance, its mutations result in Fanconi anemia. (Review) PMID: 24938228
- Data shed light on SLX4 recruitment, and they point to the existence of currently unidentified ubiquitylated ligands and E3 ligases that are crucial for ICL repair. PMID: 24794496
- The interactions of SLX4 with SUMO and ubiquitin increase its affinity for factors recognizing different DNA lesions or telomeres, helping to direct the SLX4 complex in distinct functional contexts. PMID: 25533185
- The SLX4 complex is a SUMO E3 ligase that SUMOylates SLX4 itself and the XPF subunit of the DNA repair/recombination XPF-ERCC1 endonuclease. PMID: 25533188
- GEN1 activity cannot be substituted for the SLX4-associated nucleases, and one of the HJ resolvase activities, either of those associated with SLX4 or with GEN1, is required for cell viability, even in the presence of BLM. PMID: 24080495
- Most, but not all, SLX4 foci localize to telomeres in a range of human cell lines irrespective of the mechanisms used to maintain telomere length. PMID: 23994477
- SLX4 assembles an endonuclease toolkit that negatively regulates telomere length via SLX1-catalyzed nucleolytic resolution of telomere DNA structures. PMID: 24012755
- Direct interaction of Vpr with SLX4 induced the recruitment of VPRBP and kinase-active PLK1, enhancing the cleavage of DNA by SLX4-associated MUS81-EME1 endonucleases and show that the SLX4com is involved in suppressing spontaneous and HIV-1-mediated induction of type 1 interferon and establishment of antiviral responses. PMID: 24412650
- Data indicate that SLX4 mutation screening will have a very low impact (if any) in the genetic counseling of non-BRCA1/2 families. PMID: 23211700
- Data indicate that germline mutations in SLX4 are very rare and are unlikely to make a significant contribution to familial breast cancer. PMID: 22911665
- SLX4-dependent XPF-ERCC1 activity is needed for interstrand cross-linking repair. MUS81-SLX4 interaction is critical for resistance to TOP1 inhibitors.SLX4 interacts with XPF-ERCC1, MUS81-EME1, & SLX1 via MLR, SAP, & SBD domains, respectively. PMID: 23093618
- Sequencing analysis of SLX4/FANCP gene in Italian familial breast cancer cases PMID: 22383991
- Mutational analysis of SLX4 in Breast Cancer cases without mutations in BRCA1 or BRCA2 revealed extensive genetic variation. Twenty-nine novel single nucleotide variants were detected, however, none can be linked to alteration of the protein function. PMID: 22401137
- there is no evidence for a major role of SLX4 coding variants in the inherited susceptibility towards breast cancer in German and Byelorussian patients. PMID: 21805310
- biallelic mutations in SLX4 (renamed here as FANCP) cause a new subtype of Fanconi anemia, Fanconi anemia-P PMID: 21240275
- SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype PMID: 21240277
- Genetic and biochemical evidence suggest that MUS312 and BTBD12 direct Holliday junction resolution by at least two distinct endonucleases in different recombination and repair contexts. PMID: 19595722
- BTBD12/SLX4 was identified as the human ortholog of yeast DNA repair factor Slx4p and Drosophila MUS312; SLX4 assembles a modular toolkit for repair of specific types of DNA lesions and is critical for cellular responses to replication fork failure. PMID: 19596235
- Study reports the identification of Slx4 orthologs in metazoa, including fly MUS312, essential for meiotic recombination, and human BTBD12, an ATM/ATR checkpoint kinase. PMID: 19596236
- show that SLX4 binds the XPF(ERCC4) and MUS81 subunits of the XPF-ERCC1 and MUS81-EME1 endonucleases and is required for DNA interstrand crosslink repair. PMID: 19596236
显示更多
收起更多
-
相关疾病:Fanconi anemia complementation group P (FANCP)
-
亚细胞定位:Nucleus. Note=Localizes to sites of DNA damage.
-
蛋白家族:SLX4 family
-
数据库链接:
Most popular with customers
-
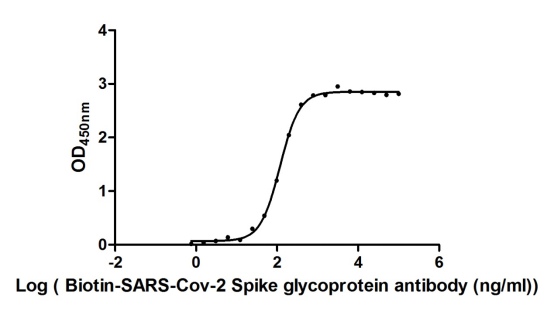
Recombinant Severe acute respiratory syndrome coronavirus 2 Spike glycoprotein (S), partial (Active)
Express system: Mammalian cell
Species: Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) (SARS-CoV-2)
-
Recombinant Human Cytokine receptor common subunit beta (CSF2RB), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Angiopoietin-2 (ANGPT2) (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Claudin-6 (CLDN6)-VLPs, Fluorescent (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
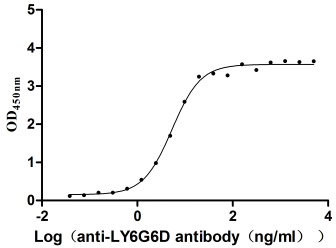
Recombinant Macaca fascicularis lymphocyte antigen 6 family member G6D (LY6G6D) (Active)
Express system: Yeast
Species: Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
-
Recombinant Human Urokinase-type plasminogen activator(PLAU) (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
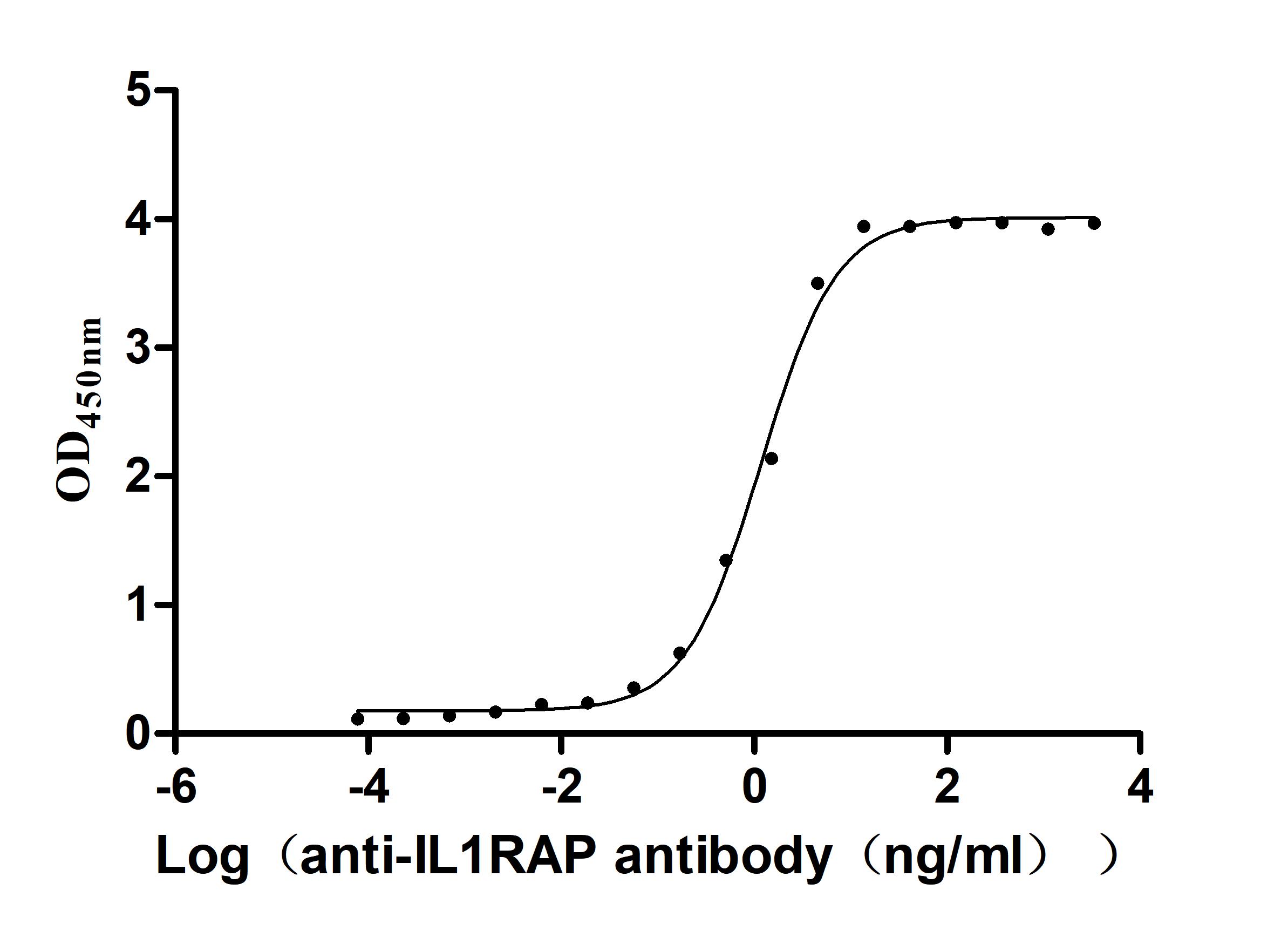
Recombinant Human Interleukin-1 receptor accessory protein (IL1RAP), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
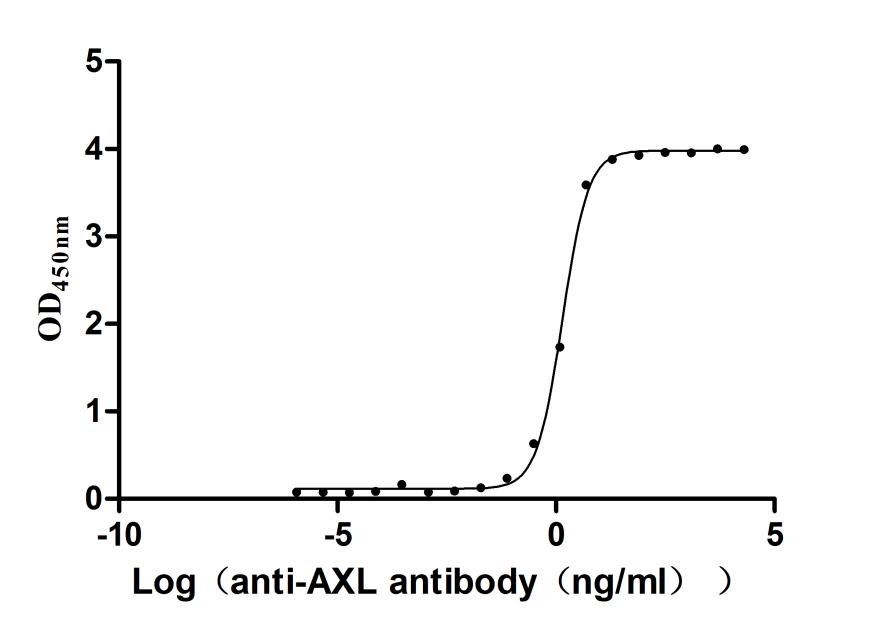
Recombinant Human Tyrosine-protein kinase receptor UFO(AXL),partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)


-AC1.jpg)
f4-AC1.jpg)