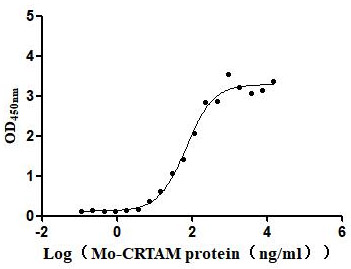
Recombinant Escherichia coli Ribonuclease E (rne), partial
-
中文名称:Recombinant Escherichia coli Ribonuclease E (rne), partial
-
货号:CSB-YP324207ENV
-
规格:¥2616
-
图片:
-
其他:
产品详情
-
纯度:Greater than 95% as determined by SDS-PAGE.
-
生物活性:Not Test
-
基因名:rne
-
Uniprot No.:
-
别名:RNase E
-
种属:Escherichia coli (strain K12)
-
蛋白长度:Partial
-
来源:Yeast
-
分子量:13.9 kDa
-
表达区域:35-125aa
-
氨基酸序列EQKKANIYKGKITRIEPSLEAAFVDYGAERHGFLPLKEIAREYFPANYSAHGRPNIKDVLREGQEVIVQIDKEERGNKGAALTTFISLAGS
Note: The complete sequence including tag sequence, target protein sequence and linker sequence could be provided upon request. -
蛋白标签:C-terminal 6xHis-Myc-tagged
-
产品提供形式:Liquid or Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
缓冲液:If the delivery form is liquid, the default storage buffer is Tris/PBS-based buffer, 5%-50% glycerol. If the delivery form is lyophilized powder, the buffer before lyophilization is Tris/PBS-based buffer, 6% Trehalose, pH 8.0.
-
复溶:We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20℃/-80℃. Our default final concentration of glycerol is 50%. Customers could use it as reference.
-
储存条件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保质期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
货期:Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.
-
注意事项:Repeated freezing and thawing is not recommended. Store working aliquots at 4℃ for up to one week.
-
Datasheet & COA:Please contact us to get it.
相关产品
靶点详情
-
功能:Endoribonuclease that plays a central role in RNA processing and decay. Required for the maturation of 5S and 16S rRNAs and the majority of tRNAs. Also involved in the degradation of most mRNAs. Can also process other RNA species, such as RNAI, a molecule that controls the replication of ColE1 plasmid, and the cell division inhibitor DicF-RNA. It initiates the decay of RNAs by cutting them internally near their 5'-end. It is able to remove poly(A) tails by an endonucleolytic process. Required to initiate rRNA degradation during both starvation and quality control; acts after RNase PH (rph) exonucleolytically digests the 3'-end of the 16S rRNA. Degradation of 16S rRNA leads to 23S rRNA degradation. Processes the 3 tRNA(Pro) precursors immediately after the 3'-CCA to generate the mature ends.; Prefers 5'-monophosphorylated substrates over 5'-triphosphorylated substrates. 5'-monophosphate-assisted cleavage requires at least 2 and preferably 3 or more unpaired 5'-terminal nucleotides. The optimal spacing between the 5' end and the scissile phosphate appears to be 8 nucleotides. Any sequence of unpaired nucleotides at the 5'-end is tolerated.
-
基因功能参考文献:
- This study suggests that, rather than its role in the processing of any one particular substrate, its distributed functions on all the different substrates (mRNA, rRNA, and tRNA) are responsible for the essentiality of RNase E in E. coli. PMID: 28167522
- Small RNAs processing and Hfq binding determinants for RNase E: RNase E cleaves any RNA fused to the 3' end. PMID: 26531825
- bacteria producing RNase E, have nutritional requirements different from those of cells supplied with only the N-terminal catalytic region of RNase E and mitigation of RNase E deficiency by overexpression of RNase G, is also affected by carbon source. PMID: 23275245
- Electrostatic potentials of were found to be conserved, highly positive, and spread over a large surface area encompassing four putative membrane-binding regions identified in the "large" domain. PMID: 22509045
- Strains harbouring rne alleles that express variants of RNase E in which 5' sensing or direct entry are inactivated, are viable. PMID: 22074454
- Increased abundance of R-loops in the rho and nusG mutants contributes to rescue the lethality associated with loss of the two RNase E cleavage pathways. PMID: 22026368
- demonstrated that CsdA, whose expression is induced by cold shock, interacts physically and functionally with RNase E PMID: 15554978
- The RNase E tetramer has two nonequivalent subunit interfaces, one of which is mediated by a single, tetrathiol-zinc complex; one or both interfaces organize the active site, which is distinct from the primary site of RNA binding. PMID: 15779893
- Observed role of RNase E in sodB mRNA turnover. PMID: 15781494
- crystal structures of the catalytic domain of RNase E as trapped allosteric intermediates with RNA substrates PMID: 16237448
- RNase E is required for induction of the glutamate-dependent acid resistance system in a RpoS-independent manner PMID: 17213667
- RNaseE and the other constituents of the RNA degradosome are components of the bacterial cytoskeleton PMID: 17242352
- This paper demonstrated that RNase E, a key endonuclease responsible for mRNA degradation in E. coli, regulates cspE transcript stability, possibly through the assembly of a degradosome. PMID: 18177308
- RNaseE and RNA helicase B play central roles in the cytoskeletal organization of the RNA degradosome PMID: 18337249
- RraA and RraB have a conserved property to directly act on RNAse E-related enzymes and inhibit their ribonucleolytic activity. PMID: 18510556
- Study shows that the arginine-rich RNA binding domain (ARRBD) of RNase E is important for the initial endoribonucleolytic cleavage of RNAI but dispensable for the endoribonucleolytic cleavages of the Rep mRNA. PMID: 19426759
显示更多
收起更多
-
亚细胞定位:Cytoplasm. Cell inner membrane; Peripheral membrane protein; Cytoplasmic side.
-
蛋白家族:RNase E/G family, RNase E subfamily
-
数据库链接:
KEGG: ecj:JW1071
STRING: 316385.ECDH10B_1155
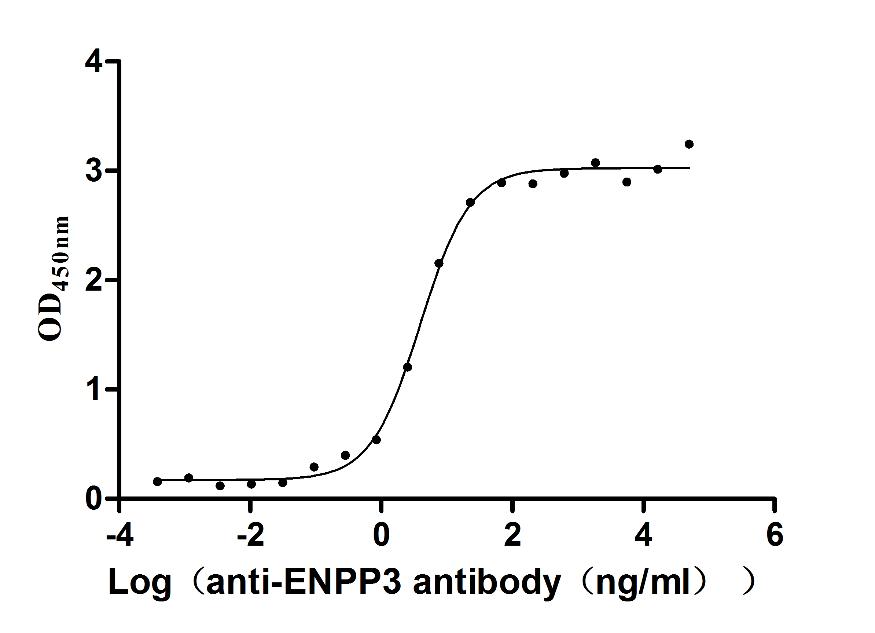
Most popular with customers
-
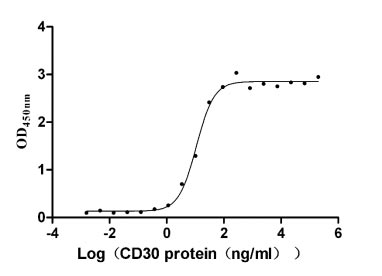
Recombinant Human Tumor necrosis factor ligand superfamily member 8 (TNFSF8), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
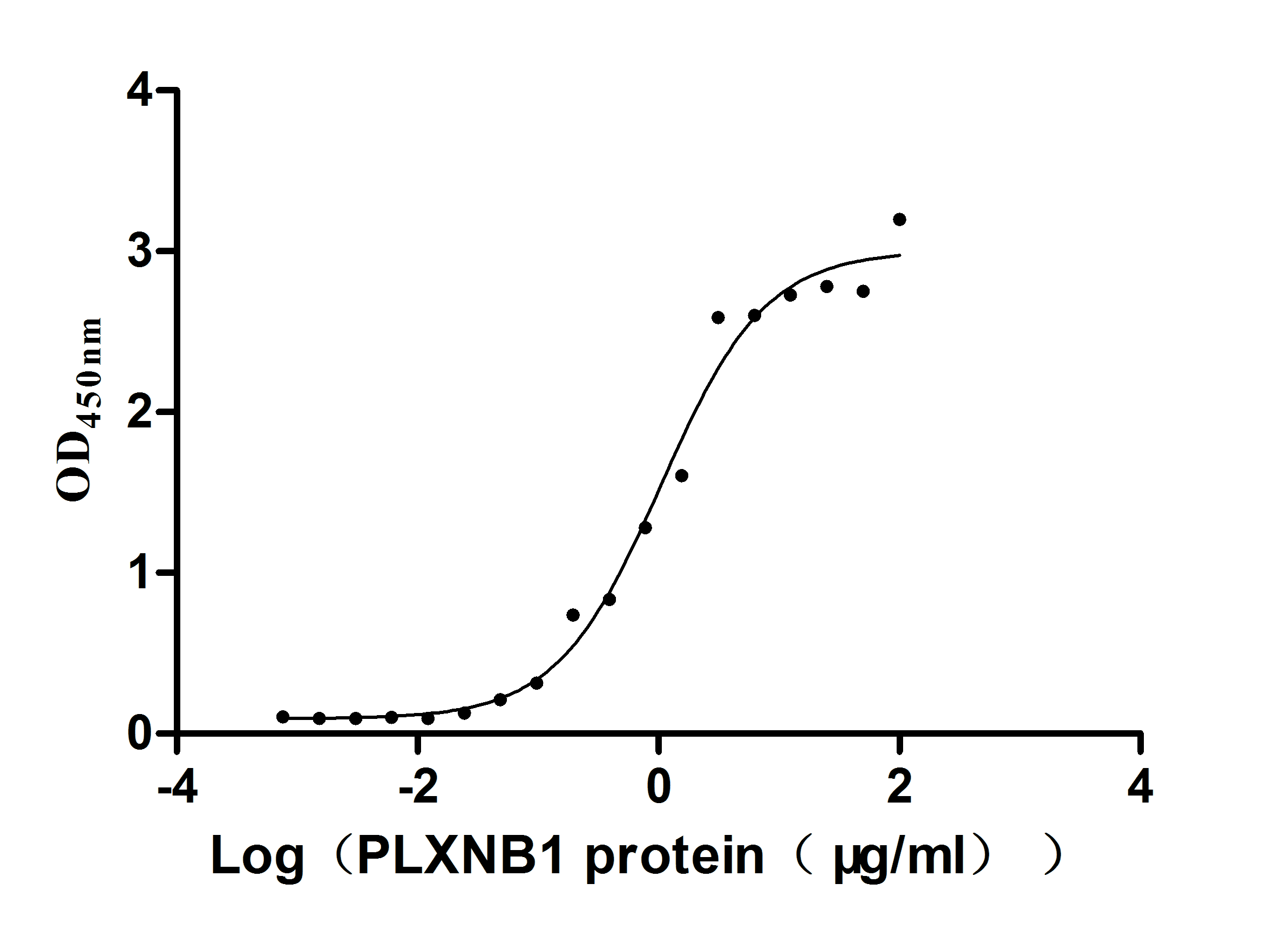
Recombinant Human Plexin-B1 (PLXNB1), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
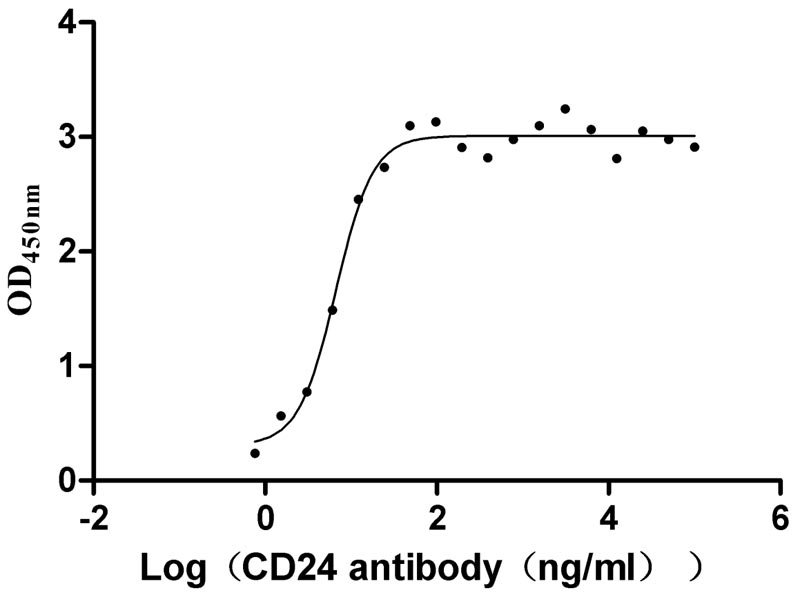
Recombinant Human Signal transducer CD24 (CD24)-Nanoparticle (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Express system: Mammalian cell
Species: Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
-
Recombinant Human Interleukin-17A (IL17A) (T26A) (Active)
Express system: Baculovirus
Species: Homo sapiens (Human)
-
Recombinant Mouse Cell adhesion molecule 1 (Cadm1), partial (Active)
Express system: Mammalian cell
Species: Mus musculus (Mouse)
-
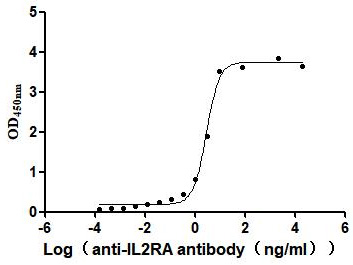
Recombinant Human Interleukin-2 receptor subunit alpha (IL2RA), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
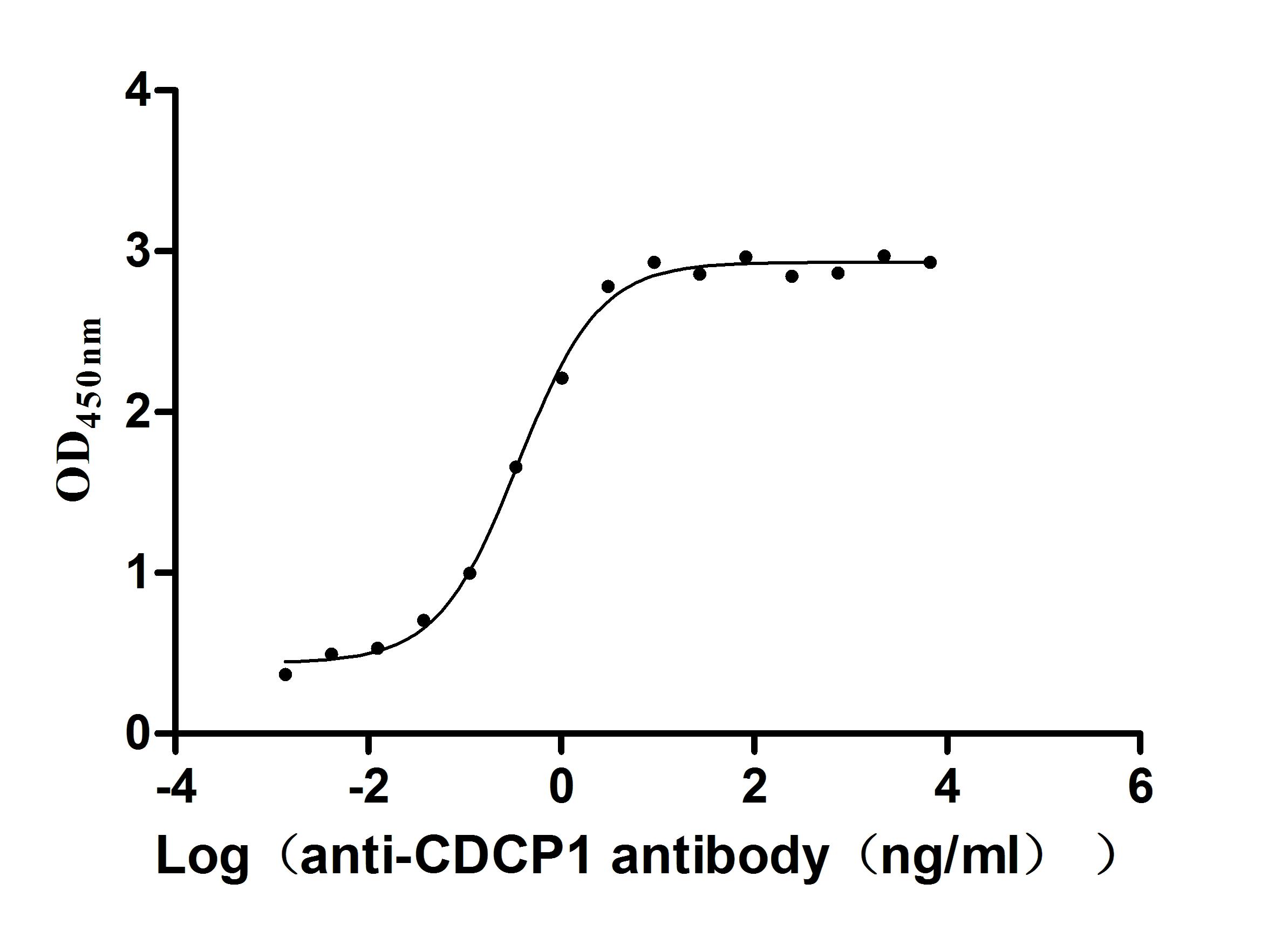
Recombinant Human CUB domain-containing protein 1 (CDCP1), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)



-AC1.jpg)