Recombinant Escherichia coli Exodeoxyribonuclease V beta chain (recB), partial
-
中文名称:大肠杆菌recB重组蛋白
-
货号:CSB-YP357378ENV
-
规格:
-
来源:Yeast
-
其他:
-
中文名称:大肠杆菌recB重组蛋白
-
货号:CSB-EP357378ENV
-
规格:
-
来源:E.coli
-
其他:
-
中文名称:大肠杆菌recB重组蛋白
-
货号:CSB-EP357378ENV-B
-
规格:
-
来源:E.coli
-
共轭:Avi-tag Biotinylated
E. coli biotin ligase (BirA) is highly specific in covalently attaching biotin to the 15 amino acid AviTag peptide. This recombinant protein was biotinylated in vivo by AviTag-BirA technology, which method is BriA catalyzes amide linkage between the biotin and the specific lysine of the AviTag.
-
其他:
-
中文名称:大肠杆菌recB重组蛋白
-
货号:CSB-BP357378ENV
-
规格:
-
来源:Baculovirus
-
其他:
-
中文名称:大肠杆菌recB重组蛋白
-
货号:CSB-MP357378ENV
-
规格:
-
来源:Mammalian cell
-
其他:
产品详情
-
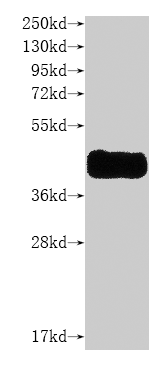
纯度:>85% (SDS-PAGE)
-
基因名:recB
-
Uniprot No.:
-
别名:recB; ior; rorA; b2820; JW2788; RecBCD enzyme subunit RecB; EC 3.1.11.5; Exodeoxyribonuclease V 135 kDa polypeptide; Exodeoxyribonuclease V beta chain; Exonuclease V subunit RecB; ExoV subunit RecB; Helicase/nuclease RecBCD subunit RecB
-
种属:Escherichia coli (strain K12)
-
蛋白长度:Partial
-
蛋白标签:Tag type will be determined during the manufacturing process.
The tag type will be determined during production process. If you have specified tag type, please tell us and we will develop the specified tag preferentially. -
产品提供形式:Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
复溶:We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20℃/-80℃. Our default final concentration of glycerol is 50%. Customers could use it as reference.
-
储存条件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保质期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
货期:Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.Note: All of our proteins are default shipped with normal blue ice packs, if you request to ship with dry ice, please communicate with us in advance and extra fees will be charged.
-
注意事项:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet :Please contact us to get it.
相关产品
靶点详情
-
功能:A helicase/nuclease that prepares dsDNA breaks (DSB) for recombinational DNA repair. Binds to DSBs and unwinds DNA via a rapid (>1 kb/second) and highly processive (>30 kb) ATP-dependent bidirectional helicase. Unwinds dsDNA until it encounters a Chi (crossover hotspot instigator, 5'-GCTGGTGG-3') sequence from the 3' direction. Cuts ssDNA a few nucleotides 3' to Chi site, by nicking one strand or switching the strand degraded (depending on the reaction conditions). The properties and activities of the enzyme are changed at Chi. The Chi-altered holoenzyme produces a long 3'-ssDNA overhang which facilitates RecA-binding to the ssDNA for homologous DNA recombination and repair. Holoenzyme degrades any linearized DNA that is unable to undergo homologous recombination. In the holoenzyme this subunit contributes ATPase, 3'-5' helicase, exonuclease activity and loads RecA onto ssDNA. The RecBC complex requires the RecD subunit for nuclease activity, but can translocate along ssDNA in both directions.
-
基因功能参考文献:
- Multiple ssDNA translocases within the RecBCD complex both before and after chi ensures processive unwinding of DNA substrates required for efficient recombination events. PMID: 29032606
- This is the first occasion that RecBCD has been demonstrated to be inhibited by DNA adducts induced by cisplatin or UV. In addition, we quantified the amounts of DNA remaining after RecBCD treatment and observed that the level of inhibition was concentration and dose dependent. A DNA-targeted 9-aminoacridinecarboxamide cisplatin analogue was also found to inhibit RecBCD activity. PMID: 29129691
- Extension of the 5'-tail of the unwound duplex induces a large conformational change in the RecD subunit, that is transferred through the RecC subunit to activate the nuclease domain of the RecB subunit. PMID: 27644322
- A new role of the RecBCD complex in the processing of DNA single-strand gaps that are generated at DNA replication-blocking lesions: independently of its nuclease or helicase activities, the entire RecBCD complex is required for recombinational repair of the gap and efficient translesion synthesis. PMID: 28369478
- Sequence-dependent nanometer-scale conformational dynamics of individual RecBCD-DNA complexes has been reported. PMID: 27220465
- RecBCD is required to complete chromosomal replication. (Review) PMID: 26003632
- A mutation within RecB (Y803H) that slows the primary translocation rate of RecBC also slows the secondary translocation rate to the same extent. PMID: 22820092
- Study hypothesized that RuvAB catalyzes replication fork reversal, RecJ and XonA blunt the double-strand end, and then RecBCD loads RecA4142 onto this end to produce SOS(Con) expression. PMID: 20304994
- crystal structure of RecBCD bound to a DNA substrate PMID: 15538360
- RecBCD enzyme overproduction impairs DNA repair and homologous recombination in Escherichia coli. PMID: 15808933
- Data suggest that RecBC and RecBCD destabilize six base pairs on binding to a blunt DNA duplex end in the absence of ATP, and that a loop can form on RecBC or RecBCD binding to DNA duplexes containing a pre-formed 3'-ssDNA tail with n > or =6 nucleotides. PMID: 16126227
- Together, these results strongly indicate that RecD overproduction prevents dissociation of RecBCD enzyme from DNA substrate and thus increases its processivity. PMID: 16377056
- The kinetics of the RecBCD-catalyzed reaction with small, single-stranded oligodeoxyribonucleotide substrates under single-turnover conditions using rapid-quench flow techniques, was studied. PMID: 16887145
- Data suggest models for the molecular mechanism of Gam-mediated inhibition of RecBCD, and propose that Gam could be a mimetic of single-stranded, and perhaps also double-stranded, DNA. PMID: 17544443
- Data show that Gam inhibits the binding of RecBCD to double-stranded DNA ends, even if RecBCD is bound to DNA before its interaction with Gam. PMID: 17583735
- Data show that the RecBCD enzyme switches lead motor subunits in response to chi recognition. PMID: 18022364
- genetic requirements for SOS induction after introduction of a double-strand break (DSB) by the I-SceI endonuclease in a RecA loading deficient recB mutant (recB1080) PMID: 18445487
- Results describe the influence of DNA end structure on the mechanism of initiation of DNA unwinding by the Escherichia coli RecBCD and RecBC helicases. PMID: 18656489
显示更多
收起更多
-
蛋白家族:Helicase family, UvrD subfamily
-
数据库链接:
KEGG: ecj:JW2788
STRING: 316385.ECDH10B_2990
Most popular with customers
-
Recombinant Human CD40 ligand (CD40LG), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
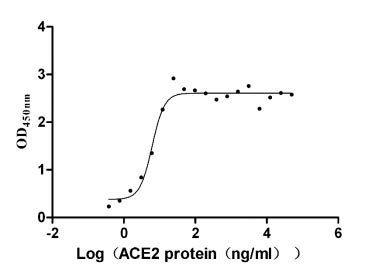
Recombinant Paguma larvata Angiotensin-converting enzyme 2 (ACE2), partial (Active)
Express system: Mammalian cell
Species: Paguma larvata (Masked palm civet)
-
Express system: Mammalian cell
Species: Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
-
Recombinant Human C-C chemokine receptor type 8 (CCR8)-VLPs (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
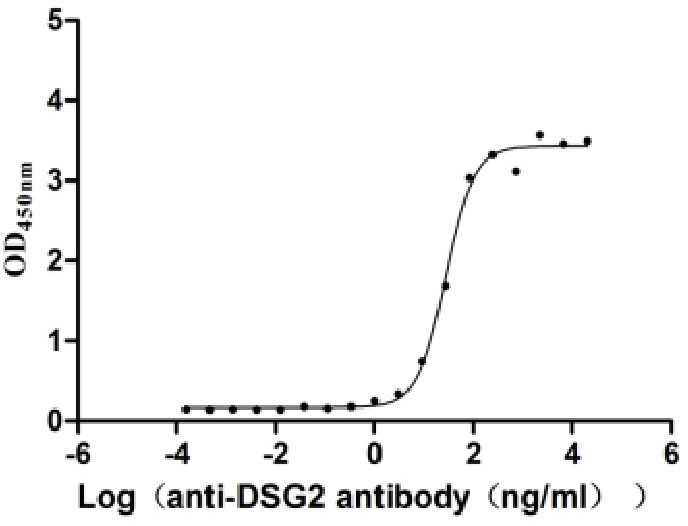
Recombinant Human Desmoglein-2 (DSG2), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Mouse CUB domain-containing protein 1 (Cdcp1), partial (Active)
Express system: Mammalian cell
Species: Mus musculus (Mouse)
-
Express system: Mammalian cell
Species: Macaca mulatta (Rhesus macaque)
-
Recombinant Human Tyrosine-protein kinase receptor UFO(AXL),partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)